PIBAdb (Polyp Image BAnk database) is a multimedia database of polyp videos and images. This database includes videos of normal colonic mucosa and colorectal polyps, both with white light and with virtual chromoscopy (Narrow Band Imaging, NBI), complemented with the corresponding definitive histological report of each polyp. The videos and images in PIBAdb are annotated manually by clinical experts participating in the PolyDeep project, which allows for the inclusion of relevant information such as the moment in time when a polyp appears on a video or its exact location on an image. PIBAdb has been suitably anonymised for use in the PolyDeep project and its subsequent transfer to the IISGS biobank (resolution of the Pontevedra-Ourense-Vigo Research Ethics Committee 2017/427). The information contained in PIBAdb may be partially or fully accessed, both for training by endoscopists in endoscopic images and for the training of automatic learning systems, as well as for other purposes, provided that they are duly justified.
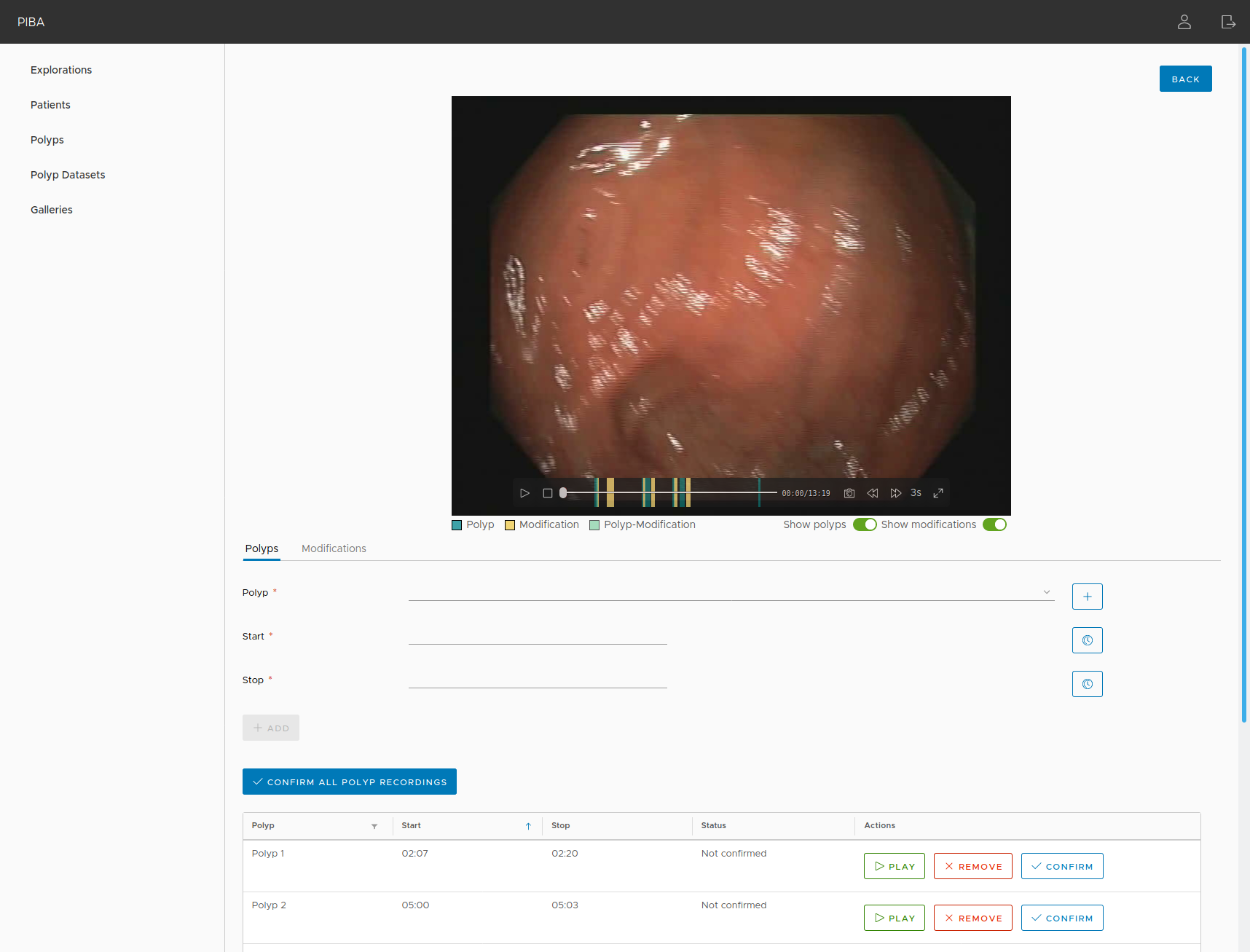
PIBA (Polyp Image BAnk) is a web application for the management of the PIBAdb database. It was developed using Angular (front-end) and Java EE (REST back-end). Users using PIBA can create upload new polyps, add histology information to them, upload videos, locate polyps in videos and images, create image galleries, and many other functionalities to explore PIBAdb or to create their own polyp database.
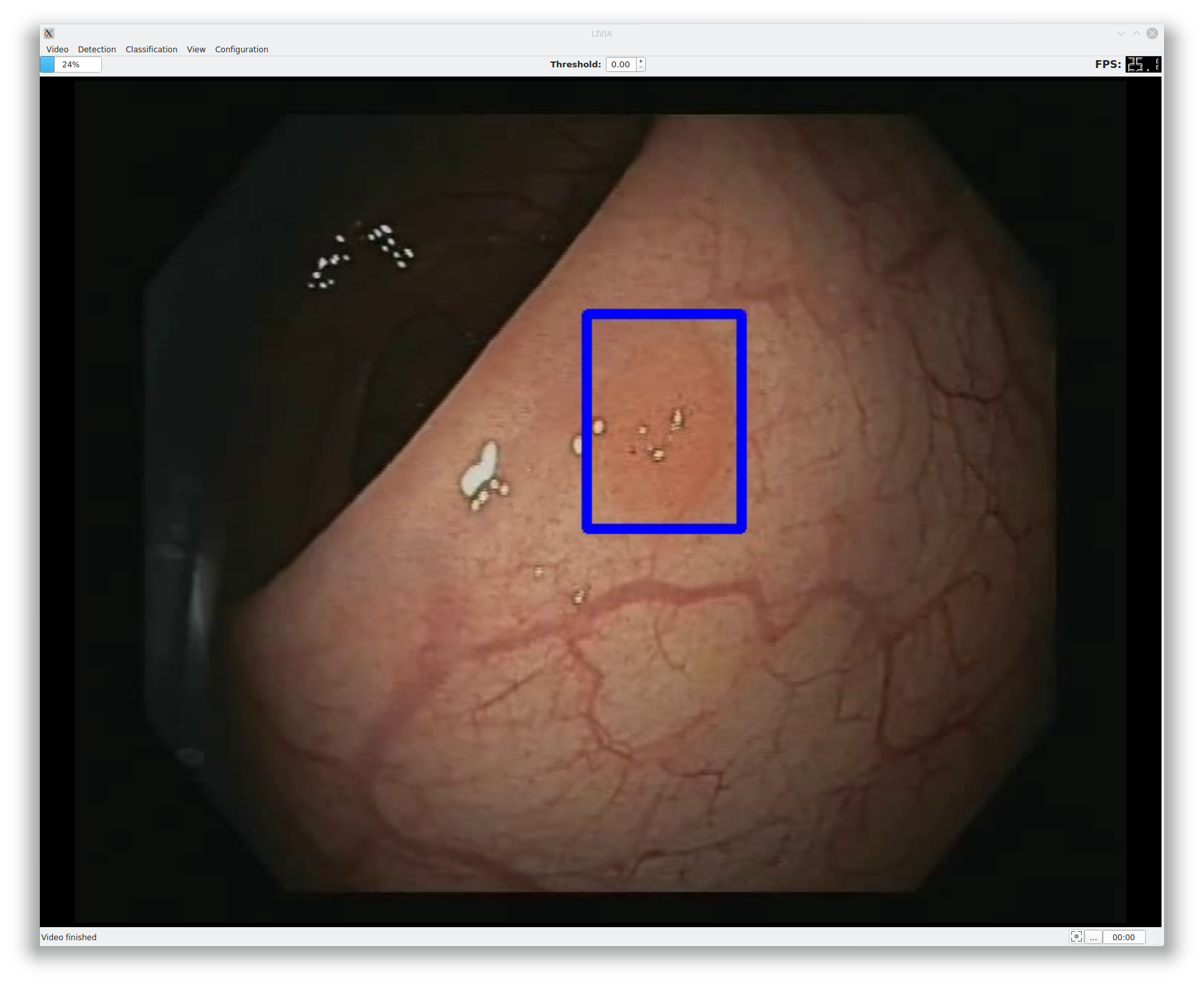
LIVIA (LIve VIdeo Analysis) is an application framework for the development of CAD (Computer-Aided Diagnosis) systems based on video. LIVIA was developed using Python, PySide2, and OpenCV. LIVIA makes it easy to create applications that show video streams and process them in real-time, modifying the video to add additional information. In this project, LIVIA was combined with Apache MxNet to detect polyps in real-time and to classify them.
Compi (Computational Pipelines) is an application framework to develop end-user, pipeline-based applications with a primary emphasis on: (i) user interface generation, by automatically generating a command-line interface based on the pipeline specific parameter definitions; (ii) application packaging, with compi-dk, which is a version-control-friendly tool to package the pipeline application and its dependencies into a Docker image; and (iii) application distribution provided through a public repository of Compi pipelines, named Compi Hub, which allows users to discover, browse and reuse them easily.

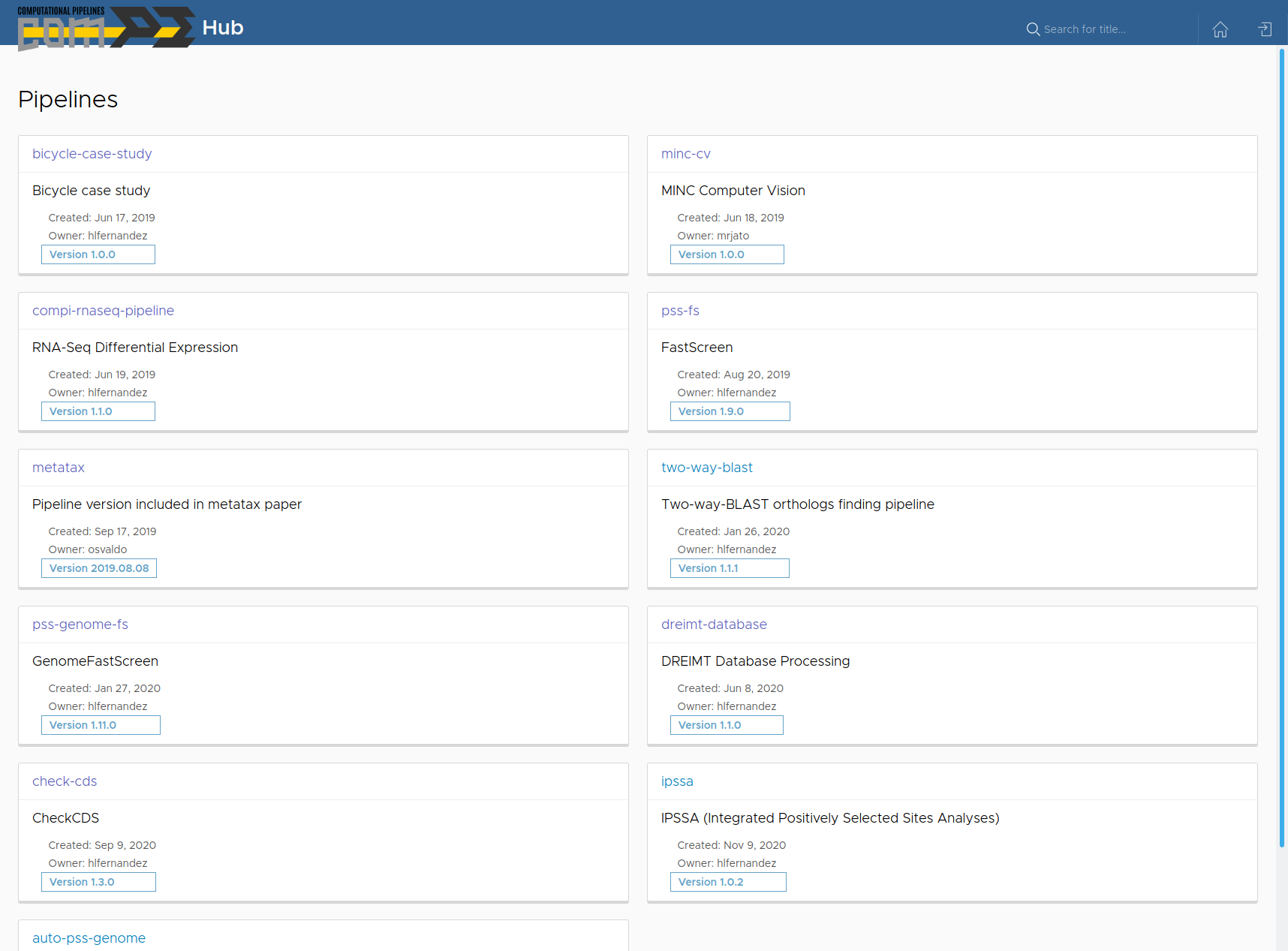
Compi Hub is a public repository of Compi pipelines that allows the community to explore them interactively. Compi Hub was developed using Angular, Node.js and MongoDB. Researchers can use Compi Hub to share their pipelines with the community or explore it to find useful pipelines shared by other researchers. Compi pipeline developers can publish their pipeline using the compi-dk tool.
Pipelines developed with Compi for the polyp detection model development and evaluation. This repository includes four pipelines for: (i) dataset split for development (training and validation datasets) and testing, (ii) model development, (iii) model evaluation (with images), and (iv) video annotation.